South Africa Hosts Data-driven Modeling Clinic
January 9, 2010
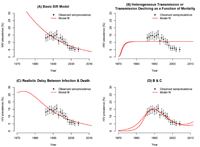
Figure 1. Six simple models of HIV transmission in Uganda. Each panel shows one or two of the models that clinic students fit to prevalence data from antenatal clinics in Uganda. Through this exercise, students were encouraged to ask meaningful epidemiological questions inspired by data: What determines peak prevalence? What factors might contribute to the decline in prevalence? What processes are necessary or sufficient to explain the patterns observed in the data? The simplest version of the transmission model (the SIR model in panel A) cannot produce a good fit to the prevalence data. By sequentially adding and removing biologically motivated details to this model, students developed an intuition for the important processes driving population-level HIV prevalence. Comparison of models illustrates that incorporating a more realistic delay between infection and death is a necessary modification to get the model to fit the data; however, this modification alone is insufficient, as shown in panel C. As in the basic SIR model, getting the model results to pass through the data requires accepting unrealistically high values for HIV prevalence in the 1970s and 1980s. Additionally incorporating either heterogeneous transmission or the impact of mortality on transmission provides an excellent fit to the data; it is not necessary to include both. After completing this iterative modeling exercise, clinic participants discussed potential methods for disentangling the influence of these two factors. Data from UNAIDS (www.unaids.org).
Many mathematicians, particularly in Africa, are highly motivated to apply their analytical skills to pressing public health problems but often have trouble bridging the gap between theory and real-world applications. In particular, they lack the skills necessary to analyze and use data in developing and testing their models. To address this need, the first Clinic on the Meaningful Modeling of Epidemiological Data was held at the African Institute for Mathematical Sciences (AIMS), in Muizenburg, South Africa, May 11�19, 2009. The clinic, which is to be held annually, brought together mathematicians, statisticians, ecologists, and epidemiologists at different stages in their careers to consider meaningful questions about infectious disease dynamics by integrating mathematical models with epidemiological data.
The clinic was sponsored and run by the Center for Discrete Mathematics and Theoretical Computer Science at Rutgers University and the Mathematical Biosciences Institute at Ohio State University, in collaboration with the South African Centre for Epidemiological Modelling and Analysis (SACEMA) and AIMS, as part of the African Biomathematics Initiative.
Techniques for linking models and data were the primary focus of the clinic. The program, a series of interactive lectures and computer tutorials, moved gradually from canned exercises to independent exploration of novel research ideas. The overarching goals of the clinic are (1) to build capacity for meaningful disease modeling research among a burgeoning group of talented mathematicians in Africa, as well as up-and-coming mathematical scientists in the U.S. with an interest in infectious diseases, (2) to better integrate researchers across mathematical modeling, epidemiology, disease ecology, and public health, (3) to stimulate productive international collaborations between Africa and North America, and (4) to develop a pedagogical approach to disease modeling that could be replicated elsewhere. In this article we briefly describe our approach to achieving these goals.
In many ways, the first clinic was an experiment. The diverse academic backgrounds and life experiences of the participants---South African undergraduate students, American graduate students, a retired WHO official, among many others---meant that everyone had unique insights to contribute; to encourage a collaborative atmosphere, the line between organizers and other participants was intentionally blurred. Most of the participants were graduate students, pursuing master's degrees or doctorates in mathematics or epidemiology; the clinic provided an atmosphere in which they could develop their research projects, with an emphasis on linking theoretical work to data. Participants were asked to bring posters on their current or previous work, and poster sessions during the first days of the clinic encouraged both social interaction and intellectual engagement.
Participants spent the first day working through a set of exercises that illustrated the fitting of a simple mathematical model of HIV transmission to HIV prevalence data from Uganda. They were asked first to think about why the simplest possible model did not satisfactorily explain observed trends. Then, by progressively modifying the mod-el, they were able to build an understanding of the assumptions needed to produce the observed patterns (see Figure 1). On the second day, after using the same tools to explore different data sets, the students were asked to interpret differences between the original and new data sets, and present their work in progress to the group.
As the clinic moved forward, the problems posed became more complex; eventually, participants were considering active research problems. In particular, given HIV testing data from antenatal clinics in Harare, Zimbabwe, they were asked to think creatively about how the age structure in the testing data could be used to infer incidence (the rate at which new infections occur) from data on HIV prevalence (the proportion of the population that is infected). They were also asked to consider how prevalence data gathered in antenatal clinics can best be used as a surrogate for prevalence in the whole population.
At first, participants were instructed to take a simple approach to model fitting, either choosing parameters that produced the best visual fit to data or using canned optimization functions. It became clear, however, that many possessed strong skills in dynamic modeling but had little training in statistics; consequently, they did not appreciate the role of random variation in producing data. Lectures on statistics were offered to address this gap, from an introduction to probability theory to a tutorial on deriving and maximizing the likelihood of a stochastic model of disease transmission. The importance of bias in generating data was introduced in several discussions, along with logistical issues associated with the acquisition of epidemiological data in a public health setting. Although the goal of the clinic was not to provide thorough training in statistics or epidemiology, students were guided toward freely available resources and encouraged to spend time developing skills in these fields after the clinic.
The greatest challenge for the organizers was to achieve an appropriate balance between teaching technical aspects of data-driven modeling and imparting a philosophy that motivates such work. With the latter in mind, the organizers had participants spend time simply looking at data, describing what they saw and discussing potential drivers of observed patterns---based not on formal mathematical models but on their intuition and real-world knowledge. In essence, the goal was to help participants develop a knack for asking meaningful questions based on their observations and realize that once a meaningful question was formed, a model could become a tool for formulating and testing hypotheses.
Importantly, the clinic relied entirely on open-access software, which means that participants will be able to continue work on projects started at the workshop. Most of the data used were also from publicly available sources. A major goal of future clinics will be the development of an interactive online research community that will facilitate long-term international collaborations between workshop participants. By posting links to publicly available data and creating forums for their discussion and analysis, this community will also stimulate the development of new collaborations between researchers on open problems in infectious disease dynamics.
Among the important lessons to emerge from the clinic is that many African participants, despite their experience in building dynamic disease models, had taken very few university-level courses in the life sciences and clearly lacked exposure to fundamental biological concepts. An overview of infectious disease biology was not possible in the time available; however, the two-week duration planned for the 2010 clinic will allow time for introductory lessons in basic microbiology, immunology, clinical epidemiology, and other biological sciences that often underlie transmission models, along with additional focus on the development of student research projects.
While a learning process for everyone, including the organizers, the interactive pedagogical approach adopted for the 2009 clinic was in large part successful. Despite the many changes planned for future clinics, participants stated that the clinic had changed the way they thought about infectious disease modeling and would lead them to redirect their careers. The best evidence of this came during the last two days of the workshop, when the students formed a discussion group to talk about their own research agendas and their plans for moving ahead. All the students who participated in these discussions expressed the intention to address specific public health problems and were able to identify specific data that they would need to acquire to address these problems. Most of these students had thought of ways to seek out these data, including the use of connections made during the clinic.
The second Clinic on the Meaningful Modeling of Epidemiological Data will be held at AIMS, May 24 to June 4, 2010. Up-to-date information on the program and instructions for applying can be found at http://lalashan.mcmaster.ca/theobio/mmed.
Acknowledgments
The clinic was supported primarily by a grant (0829652) from the U.S. National Science Foundation to DIMACS, at Rutgers University, and by SACEMA and AIMS. JP was supported in part by the Research and Policy in Infectious Disease Dynamics (RAPIDD) Program of the Science and Technology Directorate, Department of Homeland Security, and Fogarty International Center, National Institutes of Health. The clinic organizers would like to thank DIMACS, AIMS, and SACEMA staff for logistical support and Gavin Hitchcock for invaluable feedback on the pedagogical approach and on the success of the clinic.
Juliet Pulliam is a researcher at the Fogarty International Center, National Institutes of Health. Steve Bellan is a doctoral student in environmental science, policy, and management at the University of California, Berkeley. John Hargrove is director and Brian Williams is chair of the board of trustees at the South African Centre for Epidemiological Modelling and Analysis in Stellenbosch, South Africa. Fred Roberts is director of the Center for Discrete Mathematics and Theoretical Computer Science at Rutgers University. Jonathan Dushoff is a professor in the biology department at McMaster University, in Ontario, Canada.

